VRML Converters
VRML Converter for Chemical Structures
We have implemented a WWW-accessible service which accepts chemical structures in more
than 40 established 2D and 3D chemistry exchange formats and produces a VRML scene according to
user-specified visualization parameters [2].
If the exchange formats do not contain 3D
coordinates, they will be computed automatically. The application allows the
interactive change of display style and measurement of atom distances and angles by
clicking on the atoms. These are pure client-side functionalities implemented by
automaticlly generated, embedded scripts. It is also possible
to label atoms with atom properties such as atomic number, atom symbol, polarizability
or sigma-charge.

Figure 1: VRML scene of C9H12
(ball&stick display) with measurement option for the calculation of
atom distances and angles or interactive switching of display styles.
A further feature of the application is the visualization of multi-frame files.
These chemical exchange formats contain several 3D datasets of one molecular ensemble
in different time steps. These files can be used to save molecular movements
as they occur in reaction paths or molecular dynamics.
Our application saves these movements as trajectories and displays them as
animated 3D scenes.
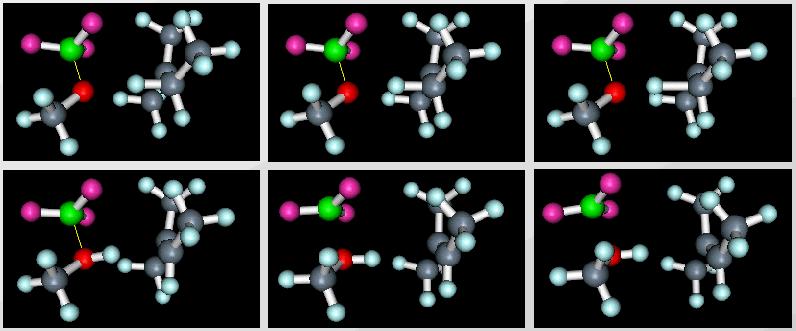
Figure 2: Snapshots of an animated VRML scene of a polybutylene reaction.
http://www2.chemie.uni-erlangen.de/services/vrmlcreator/
VRML Converter for Biological Macromolecules
Biological macromolecules, like proteins or nucleic acids (e.g. DNS) involve several 100
to several 10000 atoms. This fact leads to problems for the visualization of such
molecules. Single bonds and atoms cannot displayed anymore as separate graphical objects,
because the interactive rendering of the resulting 3D scene would be impossible on
low-budget computers. On the other side, scientists are often not interested in the
display of macromolecules which includes all the atomar detail information. Too much
information down to the level of single atoms leads to
an overloaded scene, which makes it very difficult to recognize important elements like
secondary structures. So, specific display styles for secondary structure elements have
emerged.
The visualization of these molecules requires special methods to obtain a
high rendering quality while maintaining responsiveness. We have developed an
optimizing VRML converter for biological macromolecules to their common display styles.
It allows a flexible representation of selected groups and atoms.
This option makes it
possible to add detailed atomic information into the scene only where it is useful and
required.
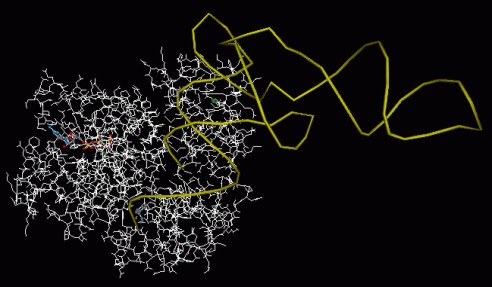
Figure 3: VRML scene of the ternary complex - elongation factor Tu,
aminoacylated tRNA and guanosinetriphosphate - of Escherichia coli.
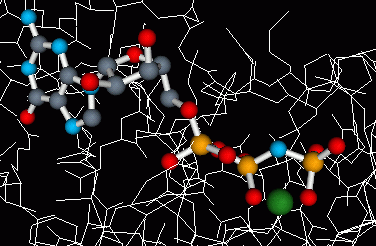
Figure 4: Guanosinetriphosphate (GTP) and GTP binding site of the
elongation factor Tu of Escherichia coli.
|

